Rawcopy
Update 2019-10-13
Running Rawcopy is now easier than ever! Rawcopy is now available on Dockerhub (https://hub.docker.com/r/rawcopy/rawcopy). There is no need to install, just run it either via Docker or Singularity.
docker run -v "Path to .CEL-files":/input:ro \
-v "Path to output":/output \
-u `id -u`:`id -g` \
rawcopy/rawcopy:1.1 \
2
singularity run --bind "Path to .CEL-files":/input \
--bind "Path to output":/output \
docker://rawcopy/rawcopy:1.1 \
2
- The first -v/bind is the path to the .CEL-files directory
- The second -v/bind is the path to the output directory
- The number at the end specifies the number of cores to use
About
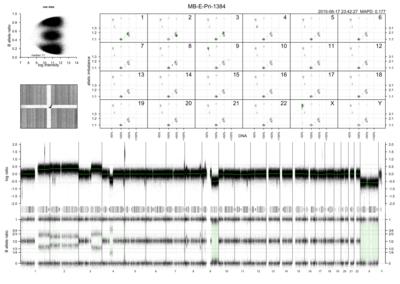
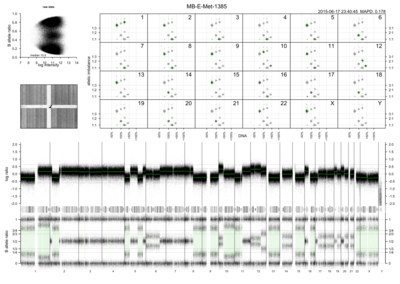
Rawcopy is an R package for processing of Affymetrix CytoScan HD, CytoScan 750k and SNP 6.0 microarrays for copy number analysis. It takes CEL files (raw microarray intensity values) as input. Output consists of:
Log ratio: normalized intensity per probe relative to sample median and a reference data set
B-allele frequency or BAF: estimated abundance of the B allele relative to total abundance, SNP probes only
Segments: genomic segments of unchanging copy number, estimated using the PSCBS package
Figures: several figures per sample and sample set are plotted for the user’s convenience, some examples are shown below
Link to article “Rawcopy: Improved copy number analysis with Affymetrix arrays”: https://www.nature.com/articles/srep36158